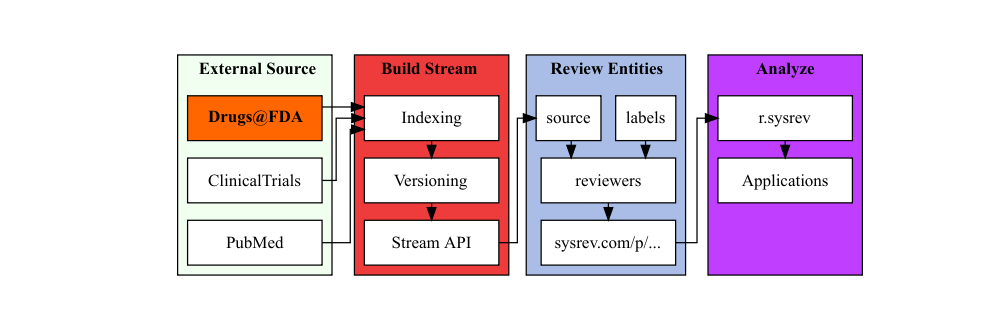
Sysrev provides a Drugs@FDA searchable stream for document reviews which politely ingests PDFs and metadata from labels.fda.gov.
The docs/metadata are indexed with a PDF parsing / OCR function to enable full text search and categorization. Sysrev also creates a versioning system for FDA documents by evaluating FDA application numbers and types.
In this vignette, you will learn to:
Search Drugs@FDA –> Review drug labels –> Analyze results.
Cardiotox@FDA sysrev
We are going to create a living review of FDA drug labels called Cardiotox@FDA.
create_sysrev("Cardiotox@FDA",get_if_exists=T)
if(interactive()){ browse_sysrev(pid=111474) }An FDA Sysrev source is created at sysrev.com/p/111474/add-articles by selecting FDA docs of application type new drug application NDA document type Review and text matching cardiotoxicity. Learn more about FDA applications types at FDA How Drugs Are Developed.
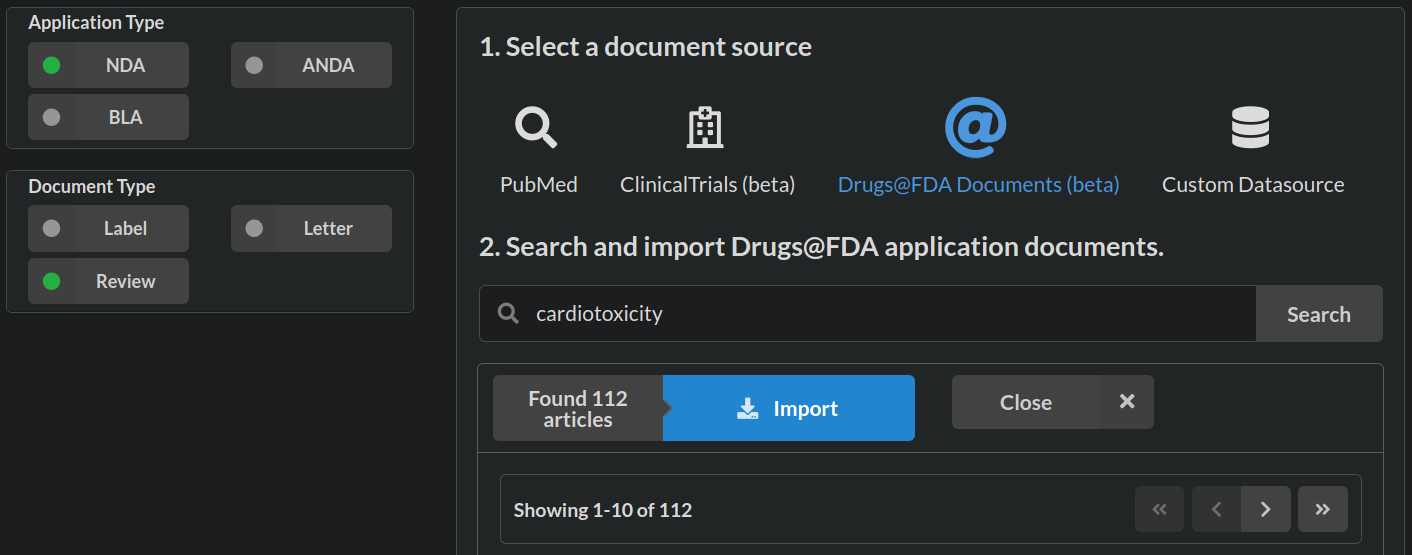
The search results in 112 articles, a manageable size for a vignette. Import pushes the labels into the Cardiotox@FDA sysrev. We can inspect the data source, add notes, and set it to automatically add new articles that match our query by clicking “check new results” at the cardiotox@fda sources page (below screenshot).
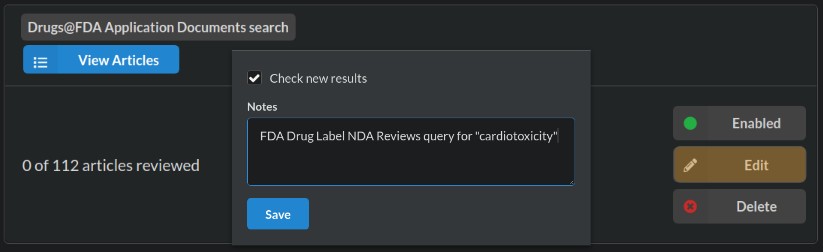
Anytime the FDA adds a new drug application review document with “cardiotoxicity” in the text it will be added to this review. Why might this be useful?
Analyzing FDA Metadata
After importing our source we can start analyzing the documents. rsr::get_article_content(111474) retrieves the article content (including meta-data) for every imported FDA drug document. The below code associates the sysrev article_id (called aid) with druglabel review type, sponsor, products data, and submission data.
meta.tb = rsr::get_article_content(111474) |> # get article content
mutate(path = glue('~/tmp/rsr/pdf/{aid}.pdf')) |> # local file names
mutate(metadata = map(metadata,jsonlite::parse_json)) |> # parse FDA meta
unnest_wider(metadata) |> # FDA meta to cols
select(aid,
fda.id = ApplNo, # FDA Application Number
sponsor = SponsorName, # Sponsor of reviewed product
review = ReviewDocumentType, # review type medical, pharma, ...
product = Products, # nested list w/ product data
submission = Submissions, # nested list w/ submission data
url = contentUrl, # a url for the included pdf
path) # a local path to store pdfs
meta.tb[1:2,]## # A tibble: 2 × 8
## aid fda.id sponsor review product submission url path
## <int> <chr> <chr> <chr> <list> <list> <chr> <glu>
## 1 13913155 207561 GILEAD SCIENCES INC medical… <list [… <list [18… https:… ~/tm…
## 2 13913084 209360 LA JOLLA PHARMA risk as… <list [… <list [1]> https:… ~/tm…Sometimes a document contains multiple submissions and products. These complex subfields can have many rows and a few columns. In the below code, we extract the active ingredient from product, and review priority / submission type from submission.
prod.tb = meta.tb |> select(aid,product) |> # build aid -> products
unnest_longer(product) |> unnest_wider(product) |> # list to columns
separate_rows(ActiveIngredient,sep="; ") |> # "name; name" -> 2 rows
select(aid, prod.active = ActiveIngredient)
sub.tb = meta.tb |> select(aid,submission) |> # aid -> submission
unnest_longer(submission) |> unnest_wider(submission) |> # list to columns
select(aid,
sub.priority = ReviewPriority,
sub.subtype = SubmissionType)Joining the meta, products and submission tables on sysrev article_id (aid) gives us a basic analyis table fda.tb. Note that article ids are repeated for documents with multiple products/submissions.
fda.tb = meta.tb |>
left_join(prod.tb,by="aid") |> left_join(sub.tb, by="aid") |>
select(aid, sponsor, review, sub.priority, sub.subtype, prod.active) |>
distinct()
fda.tb[1:2,]## # A tibble: 2 × 6
## aid sponsor review sub.priority sub.subtype prod.active
## <int> <chr> <chr> <chr> <chr> <chr>
## 1 13913155 GILEAD SCIENCES INC medical rev… STANDARD ORIG COBICISTAT
## 2 13913155 GILEAD SCIENCES INC medical rev… STANDARD SUPPL COBICISTATMeta categories
The inspectDF provides a quick way to analyze these the fda.tb.
icat <- inspectdf::inspect_cat(fda.tb)
icat |> inspectdf::show_plot(label_color = "black")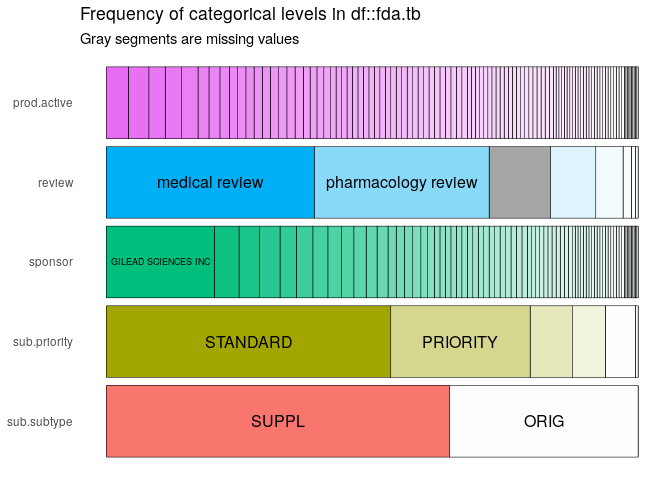
This table shows that gilead sciences has the most imported documents ( x products x submissions ). There are quite a few active ingredients, the top 5 by distinct(aid) are: COBICISTAT, SOFOSBUVIR, ELVITEGRAVIR, EMTRICITABINE, NALOXONE HYDROCHLORIDE
How many of those are Gilead active ingredients?
fda.tb |> filter(sponsor=="GILEAD SCIENCES INC") |> # GET GSI docs
count(prod.active,priority=sub.priority=="PRIORITY") |> # get count by priority
ggplot(aes(x=reorder(prod.active,n),y=n,fill=priority)) + # make a plot
geom_col(position = "stack") # + ...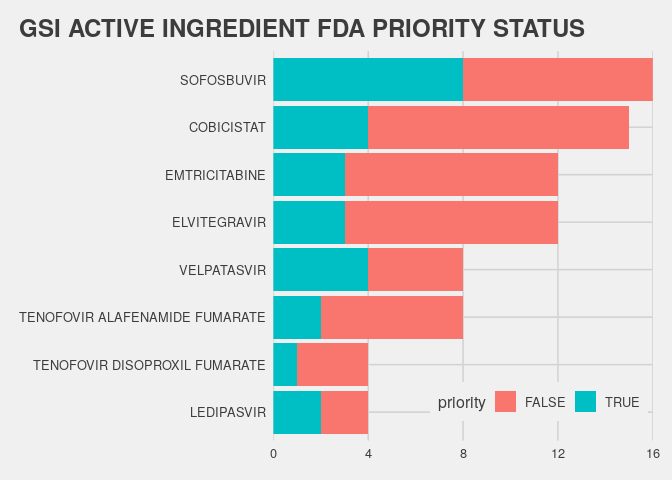
Manually Label
Sadly, most information in FDA docs is not metadata. More can be learned by reading, and extracting data from the docs. We could identify sentences that contain information like:
1. phenotypes: Where is cardiotoxicity referenced? What other phenotypes are there?
2. dose: What doses are evaluated?
3. route: How is a compound/substance delivered?
On the cardiotox@FDA labels page sysrev.com/p/111474/labels/edit we create a simple label for including articles and identifying cardiotoxicity sentences. We’ve labeled just a few articles so far. The answers can be extracted with rsr::get_answers. Other vignettes provide much more detail on the labelling process.
rsr::get_answers(111474) |>
select(aid,user_id,confirm_time,short_label,value_type,answer)## # A tibble: 4 × 6
## aid user_id confirm_time short_label value_type answer
## <int> <int> <chr> <chr> <chr> <chr>
## 1 13913147 139 2022-01-16 19:44:29 cardiotox sentence annotation "{\"34afa2…
## 2 13913147 139 2022-01-16 19:44:29 Include boolean "true"
## 3 13913153 139 2022-01-18 14:21:48 cardiotox sentence annotation "{\"a7de64…
## 4 13913153 139 2022-01-18 14:21:48 Include boolean "true"Annotating pdfs is laborious. Can we do it faster?
Automated Labels
Sysrev services provide functions that can automate document review tasks like data sourcing, extraction, review and analysis. There are two services avaiable for this project:
rsr::service_list(111474)## # A tibble: 2 × 4
## service status description reference
## <chr> <chr> <chr> <chr>
## 1 en_tox active NER for toxicology https://github.com/ontox-hu/...
## 2 export_answers active export answers to excel https://github.com/sysrev/servi…Services are still in development, but the EN_TOX tool, developed by Marc Teunis and Marie Corradi for the EU ONTOX project can automate some of the manual labelling tasks we described above. The service takes text and returns named toxicological entities. Below demonstrates how you could run this service on text from 5 imported FDA documents.
possible_text = pdftools::pdf_text |> possibly(other = NA_character_)
en_tox = partial(service_run,pid=111474,service="en_tox") |> possibly(o = tibble())
# pdf_text works pretty well
text = pdftools::pdf_text(meta.tb$path[1])
cat(text[1])
cat(text[4])
cat(text[10])
cat(text[100])
entity.fda = meta.tb |> select(aid,path) |> slice(1:5) |> # demo on 5 paths
mutate(text = pblapply(path,possible_text)) |> unnest(text) |> # pdf to text
mutate(text = gsub(pat =" +",repl = " ", trimws(text))) |> # clean text
mutate(entities = pblapply(text,en_tox)) |> # run en_tox
unnest(entities) |> # extract result
select(aid,entity,value,context) The en_tox on all pdfs is precomputed and loaded below. In a final high level analysis, entity values are counted across all the imported FDA documents. textstem::lemmatize_strings lemmatizes the values to help with the disambiguation process (i.e. human -> human or humans, cell -> cell or cells, etc.). Real disambiguation is a much bigger task.
entity.fda = readRDS(here("vignettes/data/entity.fda.RDS")) # get cached en_tox results
clean_text = compose(tolower,textstem::lemmatize_strings) # clean entity values
entity.count = entity.fda |> # lets count tox entities!
mutate(value = as.factor(clean_text(value))) |> # factors let us use forcats
select(aid,entity,value) |> distinct() |> # count entity + value articles
group_by(entity) |> # top 10 values by entity type
mutate(value = forcats::fct_lump_n(value,10)) |> # call non top 10 values 'Other'
ungroup() |>
group_by(entity,value) |>
summarize(docs = n_distinct(aid))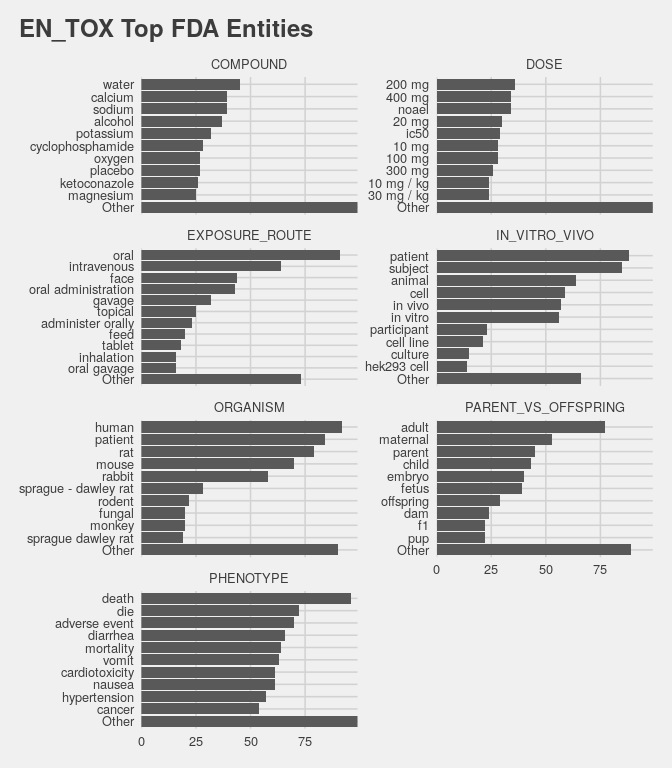
In this vignette we demonstrated integration of the Drugs@FDA resource into sysrev reviews. Sysrev services and rsr_methods were introduced:
1.rsr::create_sysrev: Create a sysrev with your sysrev token.
2.rsr::get_article_content: Get metadata and content urls from sysrev project ids.
3.rsr::get_answers: Get metadata and content urls from sysrev project ids.
4.rsr::service_run: Get a function reference to run a sysrev service.